| Product name | Evaluation slide (TEST SLIDE) |
| Cat. No. | BA |
| Current version | BA4 |
| Data sheet | BA4.pdf |
| No. of samples | 24 |
| No. of patients | 24 |
| Core diameter | 2.0 mm |
| Section thickness | 4 micrometer |
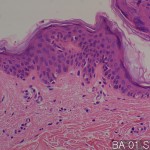
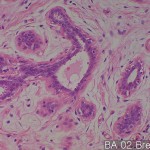
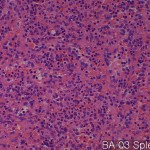
| Description | No. of normal cores: 12 cores No. or cancer cores: 12 cores The price of the TEST SLIDE is one-fourth of regular array slide. The quality of this test slide is same as the other array slides. This slide can be used for adjustment of experimental condition. The TEST SLIDE is also useful for most researchers who consider the experiment using tissue array slides. |
| Price | 177 EUR |
| 230 USD | |
| 150 GBP |
Product Related Literature
A recent study of the tissue microarray, resulting in a rapid progress in toward the quantification of a large group of biomarkers in normal and diseased tissue. However, standard procedures for the collection of tissue has not yet been established for molecular profiling.
This study presents a high-throughput analysis of non-uniformity in the breast tissue of texture image for the purpose of identifying the region of interest in the organization for molecular profiling by tissue microarray technology. Image structure breast histological slides has been described with respect to three parameters: occupancy percentage ratio of the area occupied by image block chromatin (B) as the statistical heterogeneity index (P) and such areas, the stroma , in many cases, H is used for image analysis. It uses both the color and greyscale segmentation, is defined in the thousands per image block in the data set our texture parameters are calculated. The image block, were classified into three classes by using the parameters for the new texture features statistical learning algorithm. The categories are as follows. The image block these are not unique to a specific characteristic, to block cancer tissue, the disease state and normal breast tissue to normal image block.
Lead to the identification of tissue slides the same area as the cancer-specific grayscale and color segmentation technology. In addition to the image block that has been identified as a cancer-specific cells, and is part of the area crowded them in the section of the slide image of the whole was marked by two pathologists as the unit of interest for histological study further .
After 20 years, it is true has been proven, if available, the tissue Stanford Microarray Database today, images more than 200,000 are included, these advantages, clinical diagnosis related to tissue You write, and put a cloth colored with relevant organizations metadata. The TMAD, tools for the analysis and tools import score is included via a Web interface intuitive fabric design, and image.
Adaptive segmentation of histology images in constituencies they were applied in successive stages of the two. First, we used the index of the brightness to determine the (consisting of interstitial regions abundant chromatin) the foreground from the background. For this purpose, iterative algorithm is used to adapt the two component mixture Weibull parameters for monitoring the brightness. Algorithm iterative search online to find the mixing parameters that are optimized to fit the observed values. Then, the maximum likelihood threshold value between two components is determined by the brightness threshold value for separating the interstitial rich region is stained chromatin regions.
Several models of the system and object database that are disclosed, for managing the data organization. Purpose up to the modeling of metadata for comprehensive management of tissue microarray study of a large group. There are similarities, by providing public access to the raw data fabric experiment, TMAD is different. The ongoing collaboration with research group in the United States other than remove the sample slides during the delay and resulting complications and institutions have developed transported, a clear method for transferring metadata and images from cooperative organizations As a part.
Antibodies specific for the protein used for Human Protein Atlas project administered to tissue microarrays, to create a profile of expression and localization in 48 normal human tissues, based on the systematic creation of 20 types of cancer 47 cell line published Atlas of protein antibody-based public comprehensive.